Lab 6.5.1: Subset Selection Methods#
Best Subset Selection#
# imports and setup
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from itertools import chain, combinations
import statsmodels.api as sm
pd.set_option('display.max_rows', 12)
pd.set_option('display.max_columns', 12)
pd.set_option('display.float_format', '{:20,.2f}'.format) # get rid of scientific notation
plt.style.use('seaborn') # pretty matplotlib plots
/tmp/ipykernel_3592/1468475740.py:16: MatplotlibDeprecationWarning: The seaborn styles shipped by Matplotlib are deprecated since 3.6, as they no longer correspond to the styles shipped by seaborn. However, they will remain available as 'seaborn-v0_8-<style>'. Alternatively, directly use the seaborn API instead.
plt.style.use('seaborn') # pretty matplotlib plots
hitters = pd.read_csv('../datasets/Hitters.csv', index_col=0).dropna()
hitters.index.name = 'Player'
hitters = hitters.iloc[:, [0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,19,18]] # push salary at the end
hitters['League'] = pd.get_dummies(hitters['League']).iloc[:, 0]
hitters['Division'] = pd.get_dummies(hitters['Division']).iloc[:, 0]
hitters['NewLeague'] = pd.get_dummies(hitters['NewLeague']).iloc[:, 0]
X = hitters.iloc[:, 0:19]
y = hitters.iloc[:, 19]
# adapted from:
# https://songhuiming.github.io/pages/2016/07/12/variable-selection-in-python/
# and:
# http://www.science.smith.edu/~jcrouser/SDS293/labs/
# takes a f**in' long time
k_features = 1
sub = []
reg = []
n_features = X.shape[1]
for k_features in range(1, 20):
subsets = chain(combinations(range(n_features), k_features))
best_score = np.inf
for subset in subsets:
lin_reg = sm.OLS(y, sm.add_constant(X.iloc[:, list(subset)])).fit()
score = lin_reg.ssr
if score < best_score:
best_score, best_subset = score, list(subset)
best_reg = lin_reg
sub.append(best_subset)
reg.append(best_reg)
kft.append(k_features)
results = pd.DataFrame({'kft': kft, 'sub': sub, 'reg': reg},
columns = ['sub', 'reg', 'kft']).set_index('kft')
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[3], line 20
17 best_score = np.inf
19 for subset in subsets:
---> 20 lin_reg = sm.OLS(y, sm.add_constant(X.iloc[:, list(subset)])).fit()
21 score = lin_reg.ssr
22 if score < best_score:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:922, in OLS.__init__(self, endog, exog, missing, hasconst, **kwargs)
919 msg = ("Weights are not supported in OLS and will be ignored"
920 "An exception will be raised in the next version.")
921 warnings.warn(msg, ValueWarning)
--> 922 super(OLS, self).__init__(endog, exog, missing=missing,
923 hasconst=hasconst, **kwargs)
924 if "weights" in self._init_keys:
925 self._init_keys.remove("weights")
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:748, in WLS.__init__(self, endog, exog, weights, missing, hasconst, **kwargs)
746 else:
747 weights = weights.squeeze()
--> 748 super(WLS, self).__init__(endog, exog, missing=missing,
749 weights=weights, hasconst=hasconst, **kwargs)
750 nobs = self.exog.shape[0]
751 weights = self.weights
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:202, in RegressionModel.__init__(self, endog, exog, **kwargs)
201 def __init__(self, endog, exog, **kwargs):
--> 202 super(RegressionModel, self).__init__(endog, exog, **kwargs)
203 self.pinv_wexog: Float64Array | None = None
204 self._data_attr.extend(['pinv_wexog', 'wendog', 'wexog', 'weights'])
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:270, in LikelihoodModel.__init__(self, endog, exog, **kwargs)
269 def __init__(self, endog, exog=None, **kwargs):
--> 270 super().__init__(endog, exog, **kwargs)
271 self.initialize()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:95, in Model.__init__(self, endog, exog, **kwargs)
93 missing = kwargs.pop('missing', 'none')
94 hasconst = kwargs.pop('hasconst', None)
---> 95 self.data = self._handle_data(endog, exog, missing, hasconst,
96 **kwargs)
97 self.k_constant = self.data.k_constant
98 self.exog = self.data.exog
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:135, in Model._handle_data(self, endog, exog, missing, hasconst, **kwargs)
134 def _handle_data(self, endog, exog, missing, hasconst, **kwargs):
--> 135 data = handle_data(endog, exog, missing, hasconst, **kwargs)
136 # kwargs arrays could have changed, easier to just attach here
137 for key in kwargs:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:675, in handle_data(endog, exog, missing, hasconst, **kwargs)
672 exog = np.asarray(exog)
674 klass = handle_data_class_factory(endog, exog)
--> 675 return klass(endog, exog=exog, missing=missing, hasconst=hasconst,
676 **kwargs)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:84, in ModelData.__init__(self, endog, exog, missing, hasconst, **kwargs)
82 self.orig_endog = endog
83 self.orig_exog = exog
---> 84 self.endog, self.exog = self._convert_endog_exog(endog, exog)
86 self.const_idx = None
87 self.k_constant = 0
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:509, in PandasData._convert_endog_exog(self, endog, exog)
507 exog = exog if exog is None else np.asarray(exog)
508 if endog.dtype == object or exog is not None and exog.dtype == object:
--> 509 raise ValueError("Pandas data cast to numpy dtype of object. "
510 "Check input data with np.asarray(data).")
511 return super(PandasData, self)._convert_endog_exog(endog, exog)
ValueError: Pandas data cast to numpy dtype of object. Check input data with np.asarray(data).
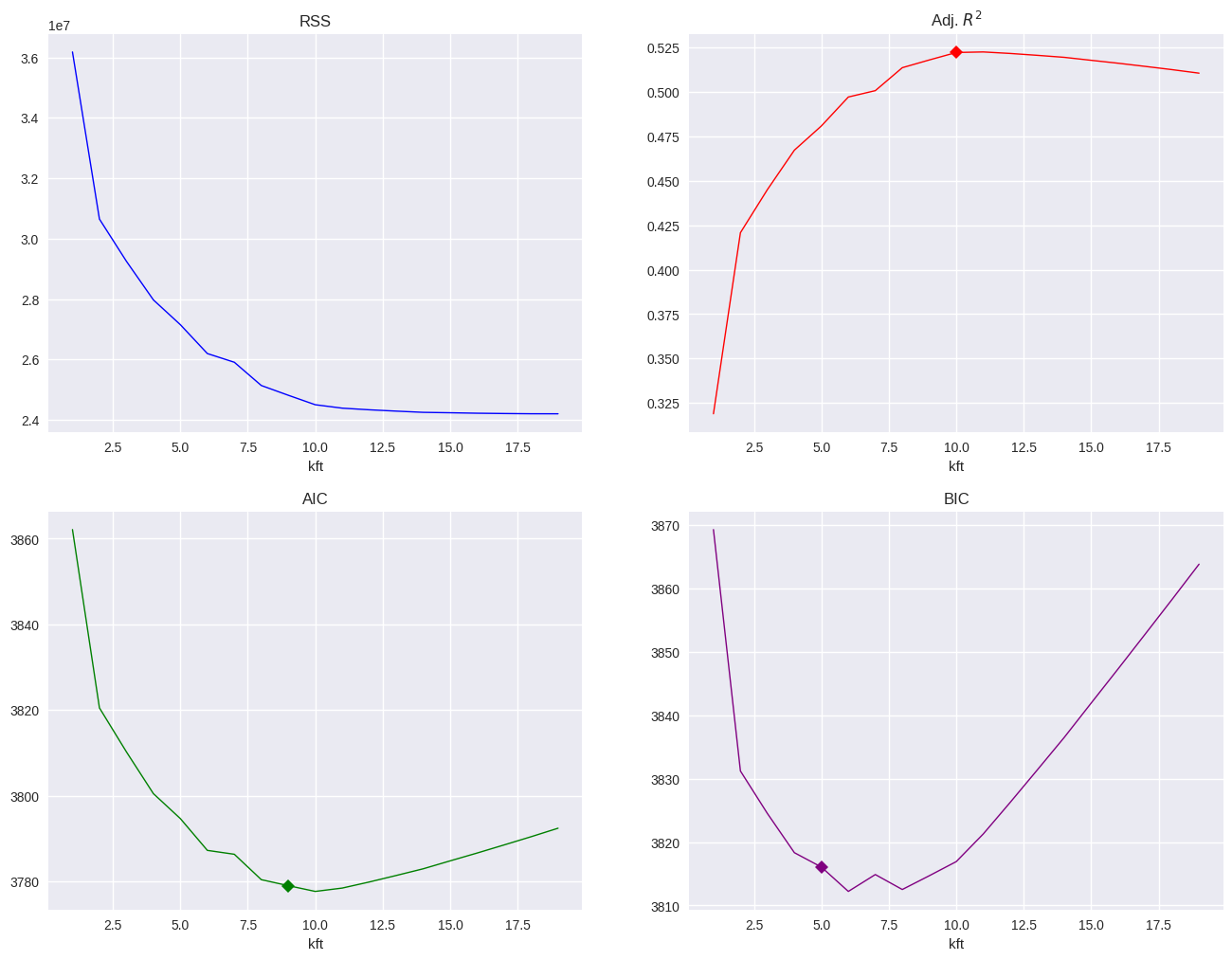
results = pd.read_pickle('results_651')
plt.figure(figsize=(16,12))
plt.subplot(2, 2, 1)
results.rss.plot(title='RSS', lw=1, color='blue')
plt.subplot(2, 2, 2)
max_r2a = results.r2a.argmax()-1
results.r2a.plot(title='Adj. $R^2$', markevery=[max_r2a], marker='D', lw=1, color='red')
plt.subplot(2, 2, 3)
min_aic = results.aic.argmin()-1
results.aic.plot(title='AIC', markevery=[min_aic], marker='D', lw=1, color='green')
plt.subplot(2, 2, 4)
min_bic = results.bic.argmin()-1
results.bic.plot(title='BIC', markevery=[min_bic], marker='D', lw=1, color='purple');
bic_reg = sm.OLS(y, sm.add_constant(X.iloc[:, [0, 1, 5, 11, 14, 15]])).fit()
bic_reg.summary()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[6], line 1
----> 1 bic_reg = sm.OLS(y, sm.add_constant(X.iloc[:, [0, 1, 5, 11, 14, 15]])).fit()
2 bic_reg.summary()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:922, in OLS.__init__(self, endog, exog, missing, hasconst, **kwargs)
919 msg = ("Weights are not supported in OLS and will be ignored"
920 "An exception will be raised in the next version.")
921 warnings.warn(msg, ValueWarning)
--> 922 super(OLS, self).__init__(endog, exog, missing=missing,
923 hasconst=hasconst, **kwargs)
924 if "weights" in self._init_keys:
925 self._init_keys.remove("weights")
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:748, in WLS.__init__(self, endog, exog, weights, missing, hasconst, **kwargs)
746 else:
747 weights = weights.squeeze()
--> 748 super(WLS, self).__init__(endog, exog, missing=missing,
749 weights=weights, hasconst=hasconst, **kwargs)
750 nobs = self.exog.shape[0]
751 weights = self.weights
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:202, in RegressionModel.__init__(self, endog, exog, **kwargs)
201 def __init__(self, endog, exog, **kwargs):
--> 202 super(RegressionModel, self).__init__(endog, exog, **kwargs)
203 self.pinv_wexog: Float64Array | None = None
204 self._data_attr.extend(['pinv_wexog', 'wendog', 'wexog', 'weights'])
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:270, in LikelihoodModel.__init__(self, endog, exog, **kwargs)
269 def __init__(self, endog, exog=None, **kwargs):
--> 270 super().__init__(endog, exog, **kwargs)
271 self.initialize()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:95, in Model.__init__(self, endog, exog, **kwargs)
93 missing = kwargs.pop('missing', 'none')
94 hasconst = kwargs.pop('hasconst', None)
---> 95 self.data = self._handle_data(endog, exog, missing, hasconst,
96 **kwargs)
97 self.k_constant = self.data.k_constant
98 self.exog = self.data.exog
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:135, in Model._handle_data(self, endog, exog, missing, hasconst, **kwargs)
134 def _handle_data(self, endog, exog, missing, hasconst, **kwargs):
--> 135 data = handle_data(endog, exog, missing, hasconst, **kwargs)
136 # kwargs arrays could have changed, easier to just attach here
137 for key in kwargs:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:675, in handle_data(endog, exog, missing, hasconst, **kwargs)
672 exog = np.asarray(exog)
674 klass = handle_data_class_factory(endog, exog)
--> 675 return klass(endog, exog=exog, missing=missing, hasconst=hasconst,
676 **kwargs)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:84, in ModelData.__init__(self, endog, exog, missing, hasconst, **kwargs)
82 self.orig_endog = endog
83 self.orig_exog = exog
---> 84 self.endog, self.exog = self._convert_endog_exog(endog, exog)
86 self.const_idx = None
87 self.k_constant = 0
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:509, in PandasData._convert_endog_exog(self, endog, exog)
507 exog = exog if exog is None else np.asarray(exog)
508 if endog.dtype == object or exog is not None and exog.dtype == object:
--> 509 raise ValueError("Pandas data cast to numpy dtype of object. "
510 "Check input data with np.asarray(data).")
511 return super(PandasData, self)._convert_endog_exog(endog, exog)
ValueError: Pandas data cast to numpy dtype of object. Check input data with np.asarray(data).
Forward and Backward Stepwise Selection#
This section is a shameless copy-paste from: http://www.science.smith.edu/~jcrouser/SDS293/labs/lab8/Lab 8 - Subset Selection in Python.pdf as they were pretty well structured. I made some small adaptations, that’s it.
def processSubset(feature_set):
# Fit model on feature_set and calculate RSS
model = sm.OLS(y,X[list(feature_set)])
regr = model.fit()
RSS = regr.ssr
return {'model': regr, 'RSS': RSS}
def forward(predictors):
# Pull out predictors we still need to process
remaining_predictors = [p for p in X.columns if p not in predictors]
results = []
for p in remaining_predictors:
results.append(processSubset(predictors+[p]))
# Wrap everything up in a nice dataframe
models = pd.DataFrame(results)
# Choose the model with the highest RSS
best_model = models.loc[models['RSS'].argmin()]
# Return the best model, along with some other useful information about the model
return best_model
def backward(predictors):
results = []
for combo in combinations(predictors, len(predictors)-1):
results.append(processSubset(combo))
# Wrap everything up in a nice dataframe
models = pd.DataFrame(results)
# Choose the model with the highest RSS
best_model = models.loc[models['RSS'].argmin()]
return best_model
Forward Selection:
models2 = pd.DataFrame(columns=['RSS', 'model'])
predictors = []
for i in range(1,len(X.columns)+1):
models2.loc[i] = forward(predictors)
predictors = models2.loc[i]['model'].model.exog_names
bic_f = []
for m in models2.model:
bic_f.append(m.bic)
np.array(bic_f).argmin()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[9], line 6
3 predictors = []
5 for i in range(1,len(X.columns)+1):
----> 6 models2.loc[i] = forward(predictors)
7 predictors = models2.loc[i]['model'].model.exog_names
9 bic_f = []
Cell In[8], line 9, in forward(predictors)
6 results = []
8 for p in remaining_predictors:
----> 9 results.append(processSubset(predictors+[p]))
11 # Wrap everything up in a nice dataframe
12 models = pd.DataFrame(results)
Cell In[7], line 3, in processSubset(feature_set)
1 def processSubset(feature_set):
2 # Fit model on feature_set and calculate RSS
----> 3 model = sm.OLS(y,X[list(feature_set)])
4 regr = model.fit()
5 RSS = regr.ssr
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:922, in OLS.__init__(self, endog, exog, missing, hasconst, **kwargs)
919 msg = ("Weights are not supported in OLS and will be ignored"
920 "An exception will be raised in the next version.")
921 warnings.warn(msg, ValueWarning)
--> 922 super(OLS, self).__init__(endog, exog, missing=missing,
923 hasconst=hasconst, **kwargs)
924 if "weights" in self._init_keys:
925 self._init_keys.remove("weights")
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:748, in WLS.__init__(self, endog, exog, weights, missing, hasconst, **kwargs)
746 else:
747 weights = weights.squeeze()
--> 748 super(WLS, self).__init__(endog, exog, missing=missing,
749 weights=weights, hasconst=hasconst, **kwargs)
750 nobs = self.exog.shape[0]
751 weights = self.weights
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:202, in RegressionModel.__init__(self, endog, exog, **kwargs)
201 def __init__(self, endog, exog, **kwargs):
--> 202 super(RegressionModel, self).__init__(endog, exog, **kwargs)
203 self.pinv_wexog: Float64Array | None = None
204 self._data_attr.extend(['pinv_wexog', 'wendog', 'wexog', 'weights'])
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:270, in LikelihoodModel.__init__(self, endog, exog, **kwargs)
269 def __init__(self, endog, exog=None, **kwargs):
--> 270 super().__init__(endog, exog, **kwargs)
271 self.initialize()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:95, in Model.__init__(self, endog, exog, **kwargs)
93 missing = kwargs.pop('missing', 'none')
94 hasconst = kwargs.pop('hasconst', None)
---> 95 self.data = self._handle_data(endog, exog, missing, hasconst,
96 **kwargs)
97 self.k_constant = self.data.k_constant
98 self.exog = self.data.exog
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:135, in Model._handle_data(self, endog, exog, missing, hasconst, **kwargs)
134 def _handle_data(self, endog, exog, missing, hasconst, **kwargs):
--> 135 data = handle_data(endog, exog, missing, hasconst, **kwargs)
136 # kwargs arrays could have changed, easier to just attach here
137 for key in kwargs:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:675, in handle_data(endog, exog, missing, hasconst, **kwargs)
672 exog = np.asarray(exog)
674 klass = handle_data_class_factory(endog, exog)
--> 675 return klass(endog, exog=exog, missing=missing, hasconst=hasconst,
676 **kwargs)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:84, in ModelData.__init__(self, endog, exog, missing, hasconst, **kwargs)
82 self.orig_endog = endog
83 self.orig_exog = exog
---> 84 self.endog, self.exog = self._convert_endog_exog(endog, exog)
86 self.const_idx = None
87 self.k_constant = 0
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:509, in PandasData._convert_endog_exog(self, endog, exog)
507 exog = exog if exog is None else np.asarray(exog)
508 if endog.dtype == object or exog is not None and exog.dtype == object:
--> 509 raise ValueError("Pandas data cast to numpy dtype of object. "
510 "Check input data with np.asarray(data).")
511 return super(PandasData, self)._convert_endog_exog(endog, exog)
ValueError: Pandas data cast to numpy dtype of object. Check input data with np.asarray(data).
Backward Selection:
models3 = pd.DataFrame(columns=['RSS', 'model'], index = range(1, len(X.columns)))
predictors = X.columns
while(len(predictors) > 1):
models3.loc[len(predictors)-1] = backward(predictors)
predictors = models3.loc[len(predictors)-1]['model'].model.exog_names
bic_b = []
for m in models3.model:
bic_b.append(m.bic)
np.array(bic_b).argmin()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[10], line 6
3 predictors = X.columns
5 while(len(predictors) > 1):
----> 6 models3.loc[len(predictors)-1] = backward(predictors)
7 predictors = models3.loc[len(predictors)-1]['model'].model.exog_names
9 bic_b = []
Cell In[8], line 25, in backward(predictors)
22 results = []
24 for combo in combinations(predictors, len(predictors)-1):
---> 25 results.append(processSubset(combo))
27 # Wrap everything up in a nice dataframe
28 models = pd.DataFrame(results)
Cell In[7], line 3, in processSubset(feature_set)
1 def processSubset(feature_set):
2 # Fit model on feature_set and calculate RSS
----> 3 model = sm.OLS(y,X[list(feature_set)])
4 regr = model.fit()
5 RSS = regr.ssr
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:922, in OLS.__init__(self, endog, exog, missing, hasconst, **kwargs)
919 msg = ("Weights are not supported in OLS and will be ignored"
920 "An exception will be raised in the next version.")
921 warnings.warn(msg, ValueWarning)
--> 922 super(OLS, self).__init__(endog, exog, missing=missing,
923 hasconst=hasconst, **kwargs)
924 if "weights" in self._init_keys:
925 self._init_keys.remove("weights")
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:748, in WLS.__init__(self, endog, exog, weights, missing, hasconst, **kwargs)
746 else:
747 weights = weights.squeeze()
--> 748 super(WLS, self).__init__(endog, exog, missing=missing,
749 weights=weights, hasconst=hasconst, **kwargs)
750 nobs = self.exog.shape[0]
751 weights = self.weights
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:202, in RegressionModel.__init__(self, endog, exog, **kwargs)
201 def __init__(self, endog, exog, **kwargs):
--> 202 super(RegressionModel, self).__init__(endog, exog, **kwargs)
203 self.pinv_wexog: Float64Array | None = None
204 self._data_attr.extend(['pinv_wexog', 'wendog', 'wexog', 'weights'])
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:270, in LikelihoodModel.__init__(self, endog, exog, **kwargs)
269 def __init__(self, endog, exog=None, **kwargs):
--> 270 super().__init__(endog, exog, **kwargs)
271 self.initialize()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:95, in Model.__init__(self, endog, exog, **kwargs)
93 missing = kwargs.pop('missing', 'none')
94 hasconst = kwargs.pop('hasconst', None)
---> 95 self.data = self._handle_data(endog, exog, missing, hasconst,
96 **kwargs)
97 self.k_constant = self.data.k_constant
98 self.exog = self.data.exog
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:135, in Model._handle_data(self, endog, exog, missing, hasconst, **kwargs)
134 def _handle_data(self, endog, exog, missing, hasconst, **kwargs):
--> 135 data = handle_data(endog, exog, missing, hasconst, **kwargs)
136 # kwargs arrays could have changed, easier to just attach here
137 for key in kwargs:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:675, in handle_data(endog, exog, missing, hasconst, **kwargs)
672 exog = np.asarray(exog)
674 klass = handle_data_class_factory(endog, exog)
--> 675 return klass(endog, exog=exog, missing=missing, hasconst=hasconst,
676 **kwargs)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:84, in ModelData.__init__(self, endog, exog, missing, hasconst, **kwargs)
82 self.orig_endog = endog
83 self.orig_exog = exog
---> 84 self.endog, self.exog = self._convert_endog_exog(endog, exog)
86 self.const_idx = None
87 self.k_constant = 0
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:509, in PandasData._convert_endog_exog(self, endog, exog)
507 exog = exog if exog is None else np.asarray(exog)
508 if endog.dtype == object or exog is not None and exog.dtype == object:
--> 509 raise ValueError("Pandas data cast to numpy dtype of object. "
510 "Check input data with np.asarray(data).")
511 return super(PandasData, self)._convert_endog_exog(endog, exog)
ValueError: Pandas data cast to numpy dtype of object. Check input data with np.asarray(data).
Choosing Among Models Using the Validation Set Approach and Cross-Validation#
Heavily borrowed from:
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5, random_state=3)
def processSubset(feature_set, X_train, y_train, X_test, y_test):
# Fit model on feature_set and calculate RSS
model = sm.OLS(y_train,X_train[list(feature_set)])
regr = model.fit()
RSS = ((regr.predict(X_test[list(feature_set)]) - y_test) ** 2).sum()
return {'model':regr, 'RSS':RSS}
def forward(predictors, X_train, y_train, X_test, y_test):
# Pull out predictors we still need to process
remaining_predictors = [p for p in X_train.columns if p not in predictors]
results = []
for p in remaining_predictors:
results.append(processSubset(predictors+[p], X_train, y_train, X_test, y_test))
# Wrap everything up in a nice dataframe
models = pd.DataFrame(results)
# Choose the model with the highest RSS
best_model = models.loc[models['RSS'].argmin()]
# Return the best model, along with some other useful information about the model
return best_model
models_train = pd.DataFrame(columns=['RSS', 'model'])
predictors = []
for i in range(1,len(X.columns)+1):
models_train.loc[i] = forward(predictors, X_train, y_train, X_test, y_test)
predictors = models_train.loc[i]['model'].model.exog_names
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[14], line 6
3 predictors = []
5 for i in range(1,len(X.columns)+1):
----> 6 models_train.loc[i] = forward(predictors, X_train, y_train, X_test, y_test)
7 predictors = models_train.loc[i]['model'].model.exog_names
Cell In[13], line 9, in forward(predictors, X_train, y_train, X_test, y_test)
6 results = []
8 for p in remaining_predictors:
----> 9 results.append(processSubset(predictors+[p], X_train, y_train, X_test, y_test))
11 # Wrap everything up in a nice dataframe
12 models = pd.DataFrame(results)
Cell In[12], line 3, in processSubset(feature_set, X_train, y_train, X_test, y_test)
1 def processSubset(feature_set, X_train, y_train, X_test, y_test):
2 # Fit model on feature_set and calculate RSS
----> 3 model = sm.OLS(y_train,X_train[list(feature_set)])
4 regr = model.fit()
5 RSS = ((regr.predict(X_test[list(feature_set)]) - y_test) ** 2).sum()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:922, in OLS.__init__(self, endog, exog, missing, hasconst, **kwargs)
919 msg = ("Weights are not supported in OLS and will be ignored"
920 "An exception will be raised in the next version.")
921 warnings.warn(msg, ValueWarning)
--> 922 super(OLS, self).__init__(endog, exog, missing=missing,
923 hasconst=hasconst, **kwargs)
924 if "weights" in self._init_keys:
925 self._init_keys.remove("weights")
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:748, in WLS.__init__(self, endog, exog, weights, missing, hasconst, **kwargs)
746 else:
747 weights = weights.squeeze()
--> 748 super(WLS, self).__init__(endog, exog, missing=missing,
749 weights=weights, hasconst=hasconst, **kwargs)
750 nobs = self.exog.shape[0]
751 weights = self.weights
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:202, in RegressionModel.__init__(self, endog, exog, **kwargs)
201 def __init__(self, endog, exog, **kwargs):
--> 202 super(RegressionModel, self).__init__(endog, exog, **kwargs)
203 self.pinv_wexog: Float64Array | None = None
204 self._data_attr.extend(['pinv_wexog', 'wendog', 'wexog', 'weights'])
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:270, in LikelihoodModel.__init__(self, endog, exog, **kwargs)
269 def __init__(self, endog, exog=None, **kwargs):
--> 270 super().__init__(endog, exog, **kwargs)
271 self.initialize()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:95, in Model.__init__(self, endog, exog, **kwargs)
93 missing = kwargs.pop('missing', 'none')
94 hasconst = kwargs.pop('hasconst', None)
---> 95 self.data = self._handle_data(endog, exog, missing, hasconst,
96 **kwargs)
97 self.k_constant = self.data.k_constant
98 self.exog = self.data.exog
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:135, in Model._handle_data(self, endog, exog, missing, hasconst, **kwargs)
134 def _handle_data(self, endog, exog, missing, hasconst, **kwargs):
--> 135 data = handle_data(endog, exog, missing, hasconst, **kwargs)
136 # kwargs arrays could have changed, easier to just attach here
137 for key in kwargs:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:675, in handle_data(endog, exog, missing, hasconst, **kwargs)
672 exog = np.asarray(exog)
674 klass = handle_data_class_factory(endog, exog)
--> 675 return klass(endog, exog=exog, missing=missing, hasconst=hasconst,
676 **kwargs)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:84, in ModelData.__init__(self, endog, exog, missing, hasconst, **kwargs)
82 self.orig_endog = endog
83 self.orig_exog = exog
---> 84 self.endog, self.exog = self._convert_endog_exog(endog, exog)
86 self.const_idx = None
87 self.k_constant = 0
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:509, in PandasData._convert_endog_exog(self, endog, exog)
507 exog = exog if exog is None else np.asarray(exog)
508 if endog.dtype == object or exog is not None and exog.dtype == object:
--> 509 raise ValueError("Pandas data cast to numpy dtype of object. "
510 "Check input data with np.asarray(data).")
511 return super(PandasData, self)._convert_endog_exog(endog, exog)
ValueError: Pandas data cast to numpy dtype of object. Check input data with np.asarray(data).
plt.plot(models_train['RSS'])
plt.xlabel('# Predictors')
plt.ylabel('RSS')
plt.plot(models_train['RSS'].argmin(), models_train['RSS'].min(), 'or');
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[15], line 4
2 plt.xlabel('# Predictors')
3 plt.ylabel('RSS')
----> 4 plt.plot(models_train['RSS'].argmin(), models_train['RSS'].min(), 'or');
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/core/base.py:742, in IndexOpsMixin.argmin(self, axis, skipna, *args, **kwargs)
738 return delegate.argmin()
739 else:
740 # error: Incompatible return value type (got "Union[int, ndarray]", expected
741 # "int")
--> 742 return nanops.nanargmin( # type: ignore[return-value]
743 delegate, skipna=skipna
744 )
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/core/nanops.py:91, in disallow.__call__.<locals>._f(*args, **kwargs)
89 if any(self.check(obj) for obj in obj_iter):
90 f_name = f.__name__.replace("nan", "")
---> 91 raise TypeError(
92 f"reduction operation '{f_name}' not allowed for this dtype"
93 )
94 try:
95 with np.errstate(invalid="ignore"):
TypeError: reduction operation 'argmin' not allowed for this dtype
models_full = pd.DataFrame(columns=['RSS', 'model'])
predictors = []
for i in range(1,20):
models_full.loc[i] = forward(predictors, X, y, X, y)
predictors = models_full.loc[i]['model'].model.exog_names
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[16], line 6
3 predictors = []
5 for i in range(1,20):
----> 6 models_full.loc[i] = forward(predictors, X, y, X, y)
7 predictors = models_full.loc[i]['model'].model.exog_names
Cell In[13], line 9, in forward(predictors, X_train, y_train, X_test, y_test)
6 results = []
8 for p in remaining_predictors:
----> 9 results.append(processSubset(predictors+[p], X_train, y_train, X_test, y_test))
11 # Wrap everything up in a nice dataframe
12 models = pd.DataFrame(results)
Cell In[12], line 3, in processSubset(feature_set, X_train, y_train, X_test, y_test)
1 def processSubset(feature_set, X_train, y_train, X_test, y_test):
2 # Fit model on feature_set and calculate RSS
----> 3 model = sm.OLS(y_train,X_train[list(feature_set)])
4 regr = model.fit()
5 RSS = ((regr.predict(X_test[list(feature_set)]) - y_test) ** 2).sum()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:922, in OLS.__init__(self, endog, exog, missing, hasconst, **kwargs)
919 msg = ("Weights are not supported in OLS and will be ignored"
920 "An exception will be raised in the next version.")
921 warnings.warn(msg, ValueWarning)
--> 922 super(OLS, self).__init__(endog, exog, missing=missing,
923 hasconst=hasconst, **kwargs)
924 if "weights" in self._init_keys:
925 self._init_keys.remove("weights")
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:748, in WLS.__init__(self, endog, exog, weights, missing, hasconst, **kwargs)
746 else:
747 weights = weights.squeeze()
--> 748 super(WLS, self).__init__(endog, exog, missing=missing,
749 weights=weights, hasconst=hasconst, **kwargs)
750 nobs = self.exog.shape[0]
751 weights = self.weights
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/regression/linear_model.py:202, in RegressionModel.__init__(self, endog, exog, **kwargs)
201 def __init__(self, endog, exog, **kwargs):
--> 202 super(RegressionModel, self).__init__(endog, exog, **kwargs)
203 self.pinv_wexog: Float64Array | None = None
204 self._data_attr.extend(['pinv_wexog', 'wendog', 'wexog', 'weights'])
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:270, in LikelihoodModel.__init__(self, endog, exog, **kwargs)
269 def __init__(self, endog, exog=None, **kwargs):
--> 270 super().__init__(endog, exog, **kwargs)
271 self.initialize()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:95, in Model.__init__(self, endog, exog, **kwargs)
93 missing = kwargs.pop('missing', 'none')
94 hasconst = kwargs.pop('hasconst', None)
---> 95 self.data = self._handle_data(endog, exog, missing, hasconst,
96 **kwargs)
97 self.k_constant = self.data.k_constant
98 self.exog = self.data.exog
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/model.py:135, in Model._handle_data(self, endog, exog, missing, hasconst, **kwargs)
134 def _handle_data(self, endog, exog, missing, hasconst, **kwargs):
--> 135 data = handle_data(endog, exog, missing, hasconst, **kwargs)
136 # kwargs arrays could have changed, easier to just attach here
137 for key in kwargs:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:675, in handle_data(endog, exog, missing, hasconst, **kwargs)
672 exog = np.asarray(exog)
674 klass = handle_data_class_factory(endog, exog)
--> 675 return klass(endog, exog=exog, missing=missing, hasconst=hasconst,
676 **kwargs)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:84, in ModelData.__init__(self, endog, exog, missing, hasconst, **kwargs)
82 self.orig_endog = endog
83 self.orig_exog = exog
---> 84 self.endog, self.exog = self._convert_endog_exog(endog, exog)
86 self.const_idx = None
87 self.k_constant = 0
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/statsmodels/base/data.py:509, in PandasData._convert_endog_exog(self, endog, exog)
507 exog = exog if exog is None else np.asarray(exog)
508 if endog.dtype == object or exog is not None and exog.dtype == object:
--> 509 raise ValueError("Pandas data cast to numpy dtype of object. "
510 "Check input data with np.asarray(data).")
511 return super(PandasData, self)._convert_endog_exog(endog, exog)
ValueError: Pandas data cast to numpy dtype of object. Check input data with np.asarray(data).
print(models_train.loc[10, 'model'].model.exog_names)
print(models_full.loc[10, 'model'].model.exog_names)
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/core/indexes/base.py:3653, in Index.get_loc(self, key)
3652 try:
-> 3653 return self._engine.get_loc(casted_key)
3654 except KeyError as err:
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/_libs/index.pyx:147, in pandas._libs.index.IndexEngine.get_loc()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/_libs/index.pyx:176, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/hashtable_class_helper.pxi:2606, in pandas._libs.hashtable.Int64HashTable.get_item()
File pandas/_libs/hashtable_class_helper.pxi:2630, in pandas._libs.hashtable.Int64HashTable.get_item()
KeyError: 10
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
Cell In[17], line 1
----> 1 print(models_train.loc[10, 'model'].model.exog_names)
2 print(models_full.loc[10, 'model'].model.exog_names)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/core/indexing.py:1096, in _LocationIndexer.__getitem__(self, key)
1094 key = tuple(com.apply_if_callable(x, self.obj) for x in key)
1095 if self._is_scalar_access(key):
-> 1096 return self.obj._get_value(*key, takeable=self._takeable)
1097 return self._getitem_tuple(key)
1098 else:
1099 # we by definition only have the 0th axis
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/core/frame.py:3877, in DataFrame._get_value(self, index, col, takeable)
3871 engine = self.index._engine
3873 if not isinstance(self.index, MultiIndex):
3874 # CategoricalIndex: Trying to use the engine fastpath may give incorrect
3875 # results if our categories are integers that dont match our codes
3876 # IntervalIndex: IntervalTree has no get_loc
-> 3877 row = self.index.get_loc(index)
3878 return series._values[row]
3880 # For MultiIndex going through engine effectively restricts us to
3881 # same-length tuples; see test_get_set_value_no_partial_indexing
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/core/indexes/base.py:3655, in Index.get_loc(self, key)
3653 return self._engine.get_loc(casted_key)
3654 except KeyError as err:
-> 3655 raise KeyError(key) from err
3656 except TypeError:
3657 # If we have a listlike key, _check_indexing_error will raise
3658 # InvalidIndexError. Otherwise we fall through and re-raise
3659 # the TypeError.
3660 self._check_indexing_error(key)
KeyError: 10
#TODO cross validation

